














 ---
### 🔍 Additional: Schema Matching with GitHub WebTables
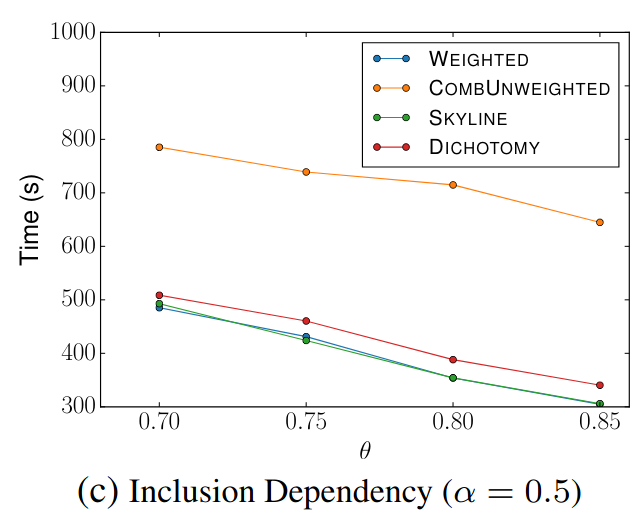
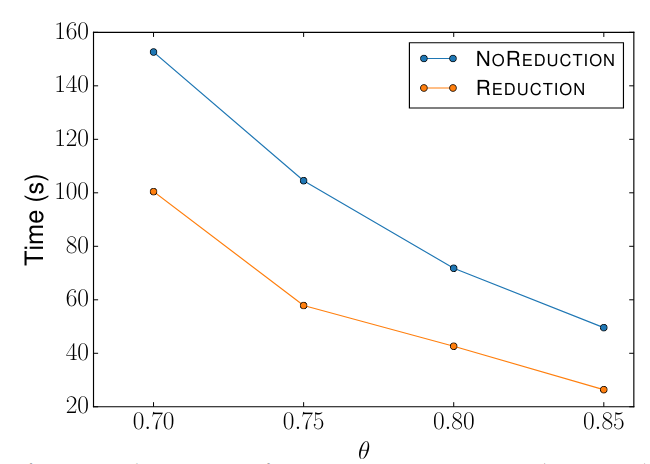
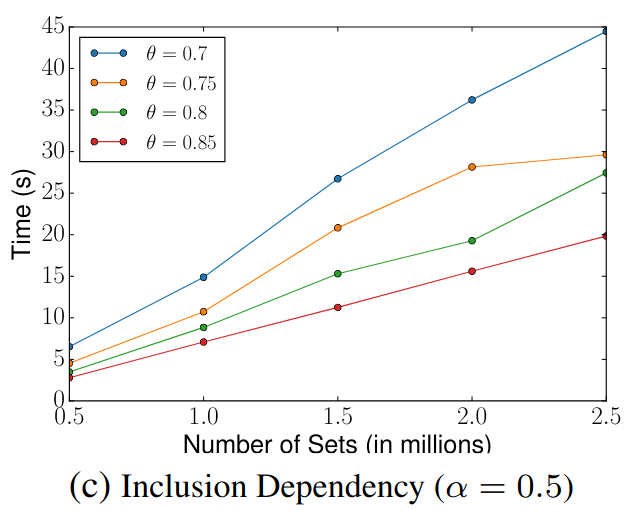
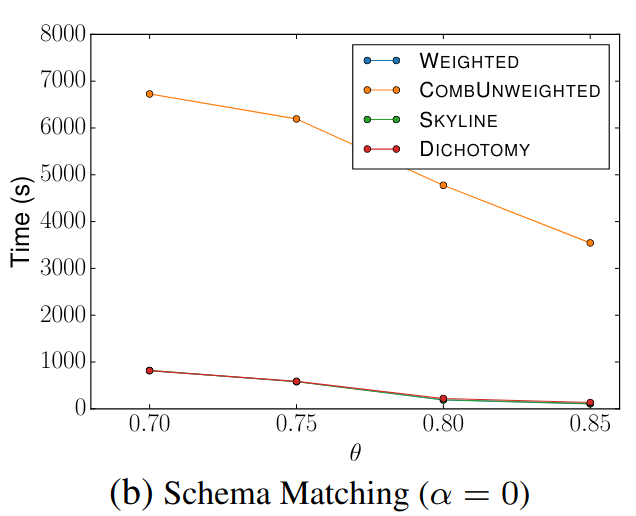
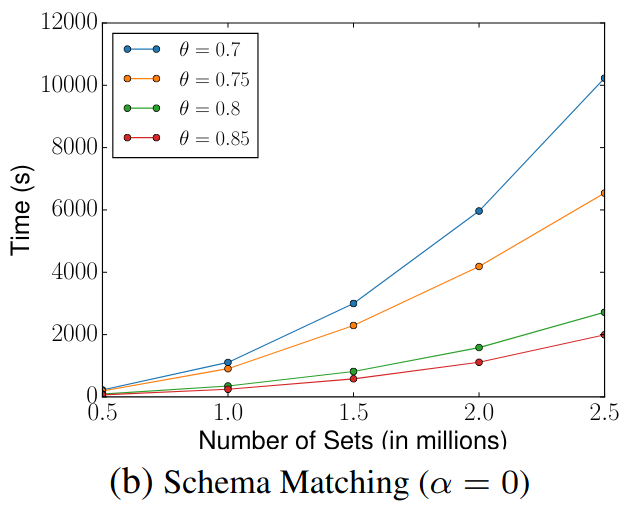
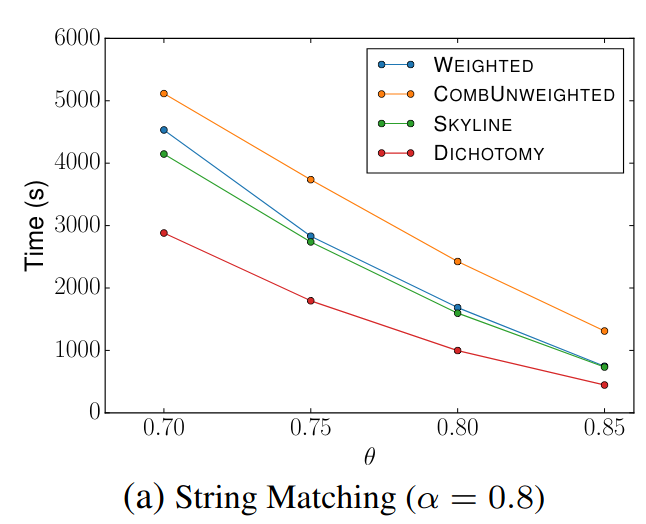
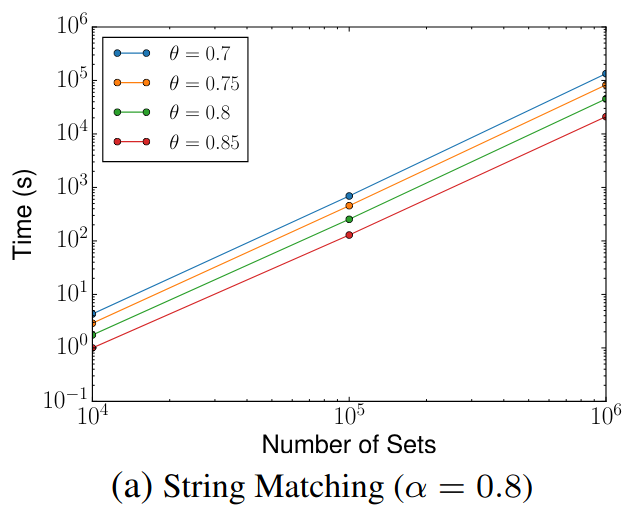
> Similar to Schema Matching, this experiment uses a GitHub WebTable as a fixed reference set and matches it against other sets. The goal is to evaluate SilkMoth’s performance across different domains.
**Left:** Matching with one reference set.
**Right:** Matching with WebTable Corpus and GitHub WebTable datasets.
The results show no significant difference, indicating consistent behavior across varying datasets.
---
### 🔍 Additional: Schema Matching with GitHub WebTables
> Similar to Schema Matching, this experiment uses a GitHub WebTable as a fixed reference set and matches it against other sets. The goal is to evaluate SilkMoth’s performance across different domains.
**Left:** Matching with one reference set.
**Right:** Matching with WebTable Corpus and GitHub WebTable datasets.
The results show no significant difference, indicating consistent behavior across varying datasets.

