🦋 LSDIPro SS2025
📄 SilkMoth: An Efficient Method for Finding Related Sets
A project inspired by the SilkMoth paper, exploring efficient techniques for related set discovery.
👥 Team Members
- Andreas Wilms
- Sarra Daknou
- Amina Iqbal
- Jakob Berschneider
📊 Experiments & Results
🧪 Interactive Demo
Follow our step-by-step Jupyter Notebook demo for a hands-on understanding of SilkMoth
📘 Project Documentation
Table of Contents
- 1. Large Scale Data Integration Project (LSDIPro)
- 2. What is SilkMoth? 🐛
- 3. The Problem 🧩
- 4. SilkMoth’s Solution 🚀
- 5. Core Pipeline Steps 🔁
- 6. Modes of Operation 🧪
- 7. Supported Similarity Functions 📐
- 8. Installing from Source
- 9. Experiment Results
1. Large Scale Data Integration Project (LSDIPro)
As part of the university project LSDIPro, our team implemented the SilkMoth paper in Python. The course focuses on large-scale data integration, where student groups reproduce and extend research prototypes.
The project emphasizes scalable algorithm design, evaluation, and handling heterogeneous data at scale.
2. What is SilkMoth?
SilkMoth is a system designed to efficiently discover related sets in large collections of data, even when the elements within those sets are only approximately similar.
This is especially important in data integration, data cleaning, and information retrieval, where messy or inconsistent data is common.
3. The Problem
Determining whether two sets are related, for example, whether two database columns should be joined, often involves comparing their elements using similarity functions (not just exact matches).
A powerful approach models this as a bipartite graph and finds the maximum matching score between elements. However, this method is computationally expensive (O(n³) per pair), making it impractical for large datasets.
4. SilkMoth’s Solution
SilkMoth tackles this with a three-step approach:
- Signature Generation: Creates compact signatures for each set, ensuring related sets share signature parts.
- Pruning: Filters out unrelated sets early, reducing candidates.
- Verification: Applies the costly matching metric only on remaining candidates, matching brute-force accuracy but faster.
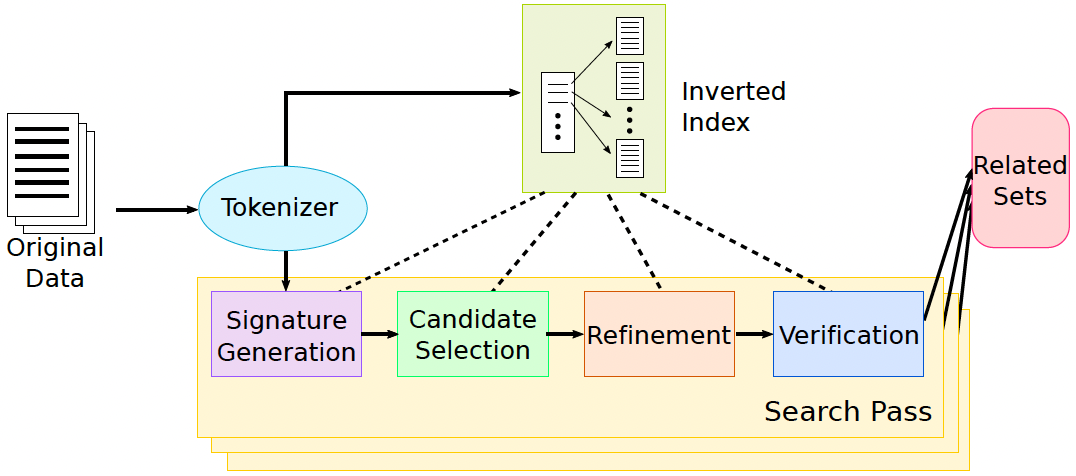
5. Core Pipeline Steps
Figure 1. SILKMOTH pipeline framework. Source: Deng et al., "SILKMOTH: An Efficient Method for Finding Related Sets with Maximum Matching Constraints", VLDB 2017. Licensed under CC BY-NC-ND 4.0.
5.1 Tokenization
Each element in every set is tokenized based on the selected similarity function:
- Jaccard Similarity: Elements are split into whitespace-delimited tokens.
- Edit Similarity: Elements are split into overlapping
q-grams (e.g., 3-grams).
5.2 Inverted Index Construction
An inverted index is built from the reference set R to map each token to a list of (set, element) pairs in which it occurs.
This allows fast lookup of candidate sets sharing tokens with a query.
5.3 Signature Generation
A signature is a subset of tokens selected from each set such that:
- Any related set must share at least one signature token.
- Signature size is minimized to reduce candidate space.
Signature selection heuristics (e.g., cost/value greedy ranking) approximate the optimal valid signature, which is NP-complete to compute exactly.
5.4 Candidate Selection
For each set R, retrieve from the inverted index all sets S sharing at least one token with R’s signature. These become candidate sets for further evaluation.
5.5 Refinement Filters
Two filters reduce false positives among candidates:
- Check Filter: Uses an upper bound on similarity to eliminate sets below threshold.
- Nearest Neighbor Filter: Approximates maximum matching score using nearest neighbor similarity for each element in
R.
5.6 Verification via Maximum Matching
Compute maximum weighted bipartite matching between elements of R and S for remaining candidates using the similarity function as edge weights.
Sets meeting or exceeding threshold δ are considered related.
6. Modes of Operation 🧪
-
Discovery Mode: Compare all pairs of sets to find all related pairs.
Use case: Finding related columns in databases. -
Search Mode: Given a reference set, find all related sets.
Use case: Schema matching or entity deduplication.
7. Supported Similarity Functions 📐
- Jaccard Similarity
- Edit Similarity (Levenshtein-based)
- Optional minimum similarity threshold
αon element comparisons.
8. Installing from Source
- Run
pip install src/to install